Computing: Difference between revisions
(Moved the screenshot) |
(→Geant3) |
||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 11: | Line 11: | ||
module unload [module] | module unload [module] | ||
To see more information and or help with this, the module man pages can be referenced either on the world wide web, or by typing <code>man module</code> | To see more information and or help with this, the module man pages can be referenced either on the world wide web, or by typing <code>man module</code> | ||
== | == ROOT == | ||
There are two versions of ROOT available to load (5.34.36 and 6.18.04) These can be loaded by issuing the following command for the respective root versions | There are two versions of ROOT available to load (5.34.36 and 6.18.04) These can be loaded by issuing the following command for the respective root versions | ||
module load root/[version] | module load root/[version] | ||
| Line 37: | Line 37: | ||
<code>docker pull sriteja/dragon_analyzer</code> will pull the Docker image. | <code>docker pull sriteja/dragon_analyzer</code> will pull the Docker image. | ||
Another issue I've faced with this is sourcing the <code>thisroot.sh</code> file at startup. So, sourcing this would have to be part of the command issued to run it. | Another issue I've faced with this is sourcing the <code>thisroot.sh</code> file at startup. So, sourcing this would have to be part of the command issued to run it when running your scripts in root. | ||
An example to demonstrate here is to convert a <code>.mid</code> file to a <code>.root</code> file. One can do this by issuing | An example to demonstrate here is to convert a <code>.mid</code> file to a <code>.root</code> file. One can do this by issuing | ||
| Line 48: | Line 48: | ||
== Geant3 == | == Geant3 == | ||
The DRAGON Geant3 simulation is a fortran based simulation package used to simulate the recoil transmission through the separator for a particular radiative capture reaction. | |||
===Branching ratio input file=== | |||
The Geant3 simulation is based on manual input for the branching ratios of the involved levels in the de-excitation of a particular resonant state following a radiative capture reaction. | |||
Information on input parameters for the .dat input file, as well as an example for the Ecm = 704 keV resonance in 22Ne(a,g)26Mg is shown below. | |||
* Input namelist for 22Ne(a,g)26Mg reaction Ecm = 557 keV | |||
* Note: All mass excesses in GeV | |||
* All widths in MeV | |||
* All elevels in MeV | |||
* beam_mass_excess and recoil_mass_excess is energy difference from (abeam or atarg)*(GeV/amu) to real value | |||
* Resonant particle's mass/energy/level defined by resenerg, not level(rstate) | |||
* Resonant mass = Beam mass + Target mass + resenergy | |||
* level(rstate) value only "names" resonant state | |||
* part_width/gam_width/spin_stat_fac/ell, define resonance width of rstate | |||
* rstate is the energy level that GEANT creates resonant particles at | |||
* level(x), specifies energy of energy level x | |||
* life(x), specifies lifetime of level(x) | |||
* Branching ratios: | |||
** br(x,z), sets branching ratio of level(x) decay mode z | |||
** md(x,z) = y, sets decay mode z, for level(x), to level(y) | |||
** example | |||
** br(2,1) = 100 | |||
** md(2,1) = 0 | |||
** sets braching ratio from level(2) to level index 1 to 100% | |||
** sets level index 1, for level(2), to level(0) or, sets branching ratio from leve(2) to level(0) to 100% | |||
$params | |||
life = 15*1000. | |||
level = 15*0. | |||
beamtyp = '22Ne' | |||
rectyp = '26Mg' | |||
zbeam = 10. | |||
abeam = 22. | |||
atarg = 4. | |||
ztarg = 2. | |||
zprod = 12. | |||
beamlifetime = 1000. | |||
beam_mass_excess = -8024.714E-6 | |||
recoil_mass_excess = -16214.546E-6 | |||
resenerg = 0.704 | |||
part_width = 0.000001 | |||
gam_width = 0.000001 | |||
spin_stat_fac = 2.0 | |||
ell = 1. | |||
rstate = 5 | |||
level( 0) = 0.0 | |||
level( 1) = 1.808 | |||
level( 2) = 2.938 | |||
level( 3) = 3.588 | |||
level( 4) = 7.061 | |||
level( 5) = 11.319 | |||
life( 0) = 1000. | |||
life( 1) = 476E-15 | |||
life( 2) = 141E-15 | |||
life( 3) = 6.45E-12 | |||
life( 4) = 7E-15 | |||
life( 5) = 1.5E-15 | |||
br(1,1) = 100. | |||
md(1,1) = 0 | |||
br(2,1) = 100. | |||
md(2,1) = 0 | |||
br(3,1) = 100. | |||
md(3,1) = 0 | |||
br(4,1) = 34.0 | |||
md(4,1) = 3 | |||
br(4,2) = 10.07 | |||
md(4,2) = 2 | |||
br(4,3) = 20.14 | |||
md(4,3) = 2 | |||
br(4,4) = 35.97 | |||
md(4,4) = 0 | |||
br(5,1) = 50.0 | |||
md(5,1) = 1 | |||
br(5,2) = 50.0 | |||
md(5,2) = 4 | |||
$[end] | |||
===.ffcards=== | |||
*The number of events is changed under: | |||
<code> TRIG 5000 </code> | |||
*Usually 5000 events provides decent statistics. | |||
*The flag for the target type (gas (0) or solid) is set via: | |||
<code> TARG 0 </code> | |||
*The lead shielding at the gas target can be included or commented out, depending on whether it was installed during the experiment: | |||
<code> SHLD 10.0 20.0 </code> | |||
===dsinit.sh=== | |||
*Before running the geant3 simulation, the dsinit.sh file needs to be updated | |||
*It is use to define the necessary environment variables for BASH shell | |||
<code> | |||
export DSROOT="`pwd`" | |||
export DSLIB="$DSROOT/lib" | |||
export DSSOURCE="$DSROOT/src" | |||
export DSINCLUDE="$DSROOT/include" | |||
export DSBIN="$DSROOT/bin" | |||
export MITRAY="$DSROOT/dragon_2014_DSSSD.dat" | |||
export FFCARD="$DSROOT/dragon_2003.ffcards" | |||
export INPUT="$DSROOT/20Nepg_397.dat" | |||
</code> | |||
*Once updated, save the file and load it by issuing the following command in the terminal: | |||
<code> source dsinit.sh </code> | |||
===Source files=== | |||
*angdist.f is used to define the gamma angular distribution | |||
After changing any of the files in the /src directory, you need to compile the simulation again via: | |||
<code> make dsinter </code> | |||
and | |||
<code> make dsbatch </code> | |||
Then copy those files one directory up, i.e., the directory where you run your simulation, and where the input files are located. | |||
===Run simulation in batch mode=== | |||
*The Geant3 simulation has 2 modes: Interactive and Batch mode | |||
*To run the simulation in batch mode issue the following command: | |||
<code> ./dsbatch </code> | |||
*Once the simulation has finished running, it creates an .hbook output file. To convert it into a .root file issue the following command: | |||
<code> h2root filename.hbook </code> | |||
== Geant4 == | == Geant4 == | ||
Coming sooner... | Coming sooner... | ||
__FORCETOC__ | __FORCETOC__ | ||
Latest revision as of 12:43, 9 November 2023
This page will detail the basics of computing with DRAGON group's servers (astro01 and astro02). The software is loaded on there through the module system.
Modules
To see the various modules/packages available to load, you can issue the following command.
module avail
You can see which modules are currently loaded by typing
module list
To load a specific module, one would use
module load [module]
Conversely, to unload a module,
module unload [module]
To see more information and or help with this, the module man pages can be referenced either on the world wide web, or by typing man module
ROOT
There are two versions of ROOT available to load (5.34.36 and 6.18.04) These can be loaded by issuing the following command for the respective root versions
module load root/[version]
Therefore, for the current modules installed, one cay type
module load root/5.34.36
and
module load root/6.18.04
To unload it, you would simply issue
module unload root/[version]
DRAGON Analyzer
DRAGON Analyzer is a software package developed for the analysis of data collected in DRAGON experiments at TRIUMF. The software is intended for experiments run with the "new" (c. 2013) DRAGON data acquisition system (VME hardware + timestamp coincidence matching).
A more comprehensive write up can be found here along with instructions to download and compile on your own machines.
However, to load the DRAGON Analyzer on astro, you may do so by issuing one of two commands:
dragon-analyzer/1.3.0_root-5.34
or
dragon-analyzer/1.3.0_root-6.18
This will load the DRAGON Analyzer and the version of ROOT it has been compiled with. This will also load the proper environmental variables needed to run the software.
Docker
Docker is a containerisation software that effectively acts as a virtual machine. In order to use this option, the user will have to have Docker or another containerisation software installed on his or her machine. Docker is not installed on astro01 or astro02. Therefore, this method is more geared towards attempting to use DRAGON Analyzer on their personal machines. If you have never used Docker, it will be helpful to familiarise yourself with some quick commands by reading through this and referring to this. Installing and setting up Docker is beyond the scope of this write up.
This is still a work in progress. One thing I am still stuck on is a way to forward X11 through. This will be updated as I have time to work on it. The Docker image is on dockerhub. However, since some of the paths are hard coded in, I would recommend building from the Dockerfile.
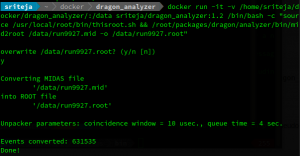
docker pull sriteja/dragon_analyzer will pull the Docker image.
Another issue I've faced with this is sourcing the thisroot.sh file at startup. So, sourcing this would have to be part of the command issued to run it when running your scripts in root.
An example to demonstrate here is to convert a .mid file to a .root file. One can do this by issuing
docker run -it -v [local directory to be volume mounted]:[target mount point] sriteja/dragon_analyzer:1.2 /bin/bash -c "/root/packages/dragon/analyzer/bin/mid2root [.mid file] -o [output root file]"
Let's dissect this:
docker run runs a specific container you mention. in this case, it is sriteja/dragon_analyzer:1.2. -it provides an interactive pseudo-tty. -v is to volume mount a directory on the container. The rest of this is simply the bash command issued. As you can see from the example, it would be prudent to call the files from and store the output files to the volume mounted location, which is /data on the container in the example.
In the case of running your analysis scripts, I've had to source thisroot.sh every time. Unless your scripts stores and outputs a file of histograms, this is not a viable method to use to run your analysis scripts [yet].
Geant3
The DRAGON Geant3 simulation is a fortran based simulation package used to simulate the recoil transmission through the separator for a particular radiative capture reaction.
Branching ratio input file
The Geant3 simulation is based on manual input for the branching ratios of the involved levels in the de-excitation of a particular resonant state following a radiative capture reaction.
Information on input parameters for the .dat input file, as well as an example for the Ecm = 704 keV resonance in 22Ne(a,g)26Mg is shown below.
- Input namelist for 22Ne(a,g)26Mg reaction Ecm = 557 keV
- Note: All mass excesses in GeV
- All widths in MeV
- All elevels in MeV
- beam_mass_excess and recoil_mass_excess is energy difference from (abeam or atarg)*(GeV/amu) to real value
- Resonant particle's mass/energy/level defined by resenerg, not level(rstate)
- Resonant mass = Beam mass + Target mass + resenergy
- level(rstate) value only "names" resonant state
- part_width/gam_width/spin_stat_fac/ell, define resonance width of rstate
- rstate is the energy level that GEANT creates resonant particles at
- level(x), specifies energy of energy level x
- life(x), specifies lifetime of level(x)
- Branching ratios:
- br(x,z), sets branching ratio of level(x) decay mode z
- md(x,z) = y, sets decay mode z, for level(x), to level(y)
- example
- br(2,1) = 100
- md(2,1) = 0
- sets braching ratio from level(2) to level index 1 to 100%
- sets level index 1, for level(2), to level(0) or, sets branching ratio from leve(2) to level(0) to 100%
$params life = 15*1000. level = 15*0. beamtyp = '22Ne' rectyp = '26Mg' zbeam = 10. abeam = 22. atarg = 4. ztarg = 2. zprod = 12. beamlifetime = 1000. beam_mass_excess = -8024.714E-6 recoil_mass_excess = -16214.546E-6 resenerg = 0.704 part_width = 0.000001 gam_width = 0.000001 spin_stat_fac = 2.0 ell = 1. rstate = 5 level( 0) = 0.0 level( 1) = 1.808 level( 2) = 2.938 level( 3) = 3.588 level( 4) = 7.061 level( 5) = 11.319 life( 0) = 1000. life( 1) = 476E-15 life( 2) = 141E-15 life( 3) = 6.45E-12 life( 4) = 7E-15 life( 5) = 1.5E-15 br(1,1) = 100. md(1,1) = 0 br(2,1) = 100. md(2,1) = 0 br(3,1) = 100. md(3,1) = 0 br(4,1) = 34.0 md(4,1) = 3 br(4,2) = 10.07 md(4,2) = 2 br(4,3) = 20.14 md(4,3) = 2 br(4,4) = 35.97 md(4,4) = 0 br(5,1) = 50.0 md(5,1) = 1 br(5,2) = 50.0 md(5,2) = 4 $[end]
.ffcards
- The number of events is changed under:
TRIG 5000
- Usually 5000 events provides decent statistics.
- The flag for the target type (gas (0) or solid) is set via:
TARG 0
- The lead shielding at the gas target can be included or commented out, depending on whether it was installed during the experiment:
SHLD 10.0 20.0
dsinit.sh
- Before running the geant3 simulation, the dsinit.sh file needs to be updated
- It is use to define the necessary environment variables for BASH shell
export DSROOT="`pwd`"
export DSLIB="$DSROOT/lib"
export DSSOURCE="$DSROOT/src"
export DSINCLUDE="$DSROOT/include"
export DSBIN="$DSROOT/bin"
export MITRAY="$DSROOT/dragon_2014_DSSSD.dat"
export FFCARD="$DSROOT/dragon_2003.ffcards"
export INPUT="$DSROOT/20Nepg_397.dat"
- Once updated, save the file and load it by issuing the following command in the terminal:
source dsinit.sh
Source files
- angdist.f is used to define the gamma angular distribution
After changing any of the files in the /src directory, you need to compile the simulation again via:
make dsinter
and
make dsbatch
Then copy those files one directory up, i.e., the directory where you run your simulation, and where the input files are located.
Run simulation in batch mode
- The Geant3 simulation has 2 modes: Interactive and Batch mode
- To run the simulation in batch mode issue the following command:
./dsbatch
- Once the simulation has finished running, it creates an .hbook output file. To convert it into a .root file issue the following command:
h2root filename.hbook
Geant4
Coming sooner...